UpSet: Gene Function Annotation!
Introduction
- An UpSet diagram is a visualization of the common and unique parts of different groups, and is an alternative to a Wayne diagram. UpSet diagrams can be used to show the common and unique parts of groups 3-7 (currently the largest setting is 7). UpSet diagrams consist of three main parts: the upper part is the number of unique and common parts of each subgroup, and the lower part is the classification of the unique and common parts of each subgroup. The left (or right) part represents the number of unduplicated elements contained in each subgroup. Each column is a grouping and each row is an element.
- In an UpSet diagram, a combination of bars and points can be used to show the intersection between individual groupings. The height of the bar graph indicates the number of elements that are unique or common to a particular grouping, while the points are used to indicate the distribution of elements across the groupings. The intersection between the groups can be visualized by the position and connection of the points.
Data from several bacteria were prepared and used to plot the upsets.
Data preparation
- All the data needed for the drawing of the UpSet plot.

Check out the details:
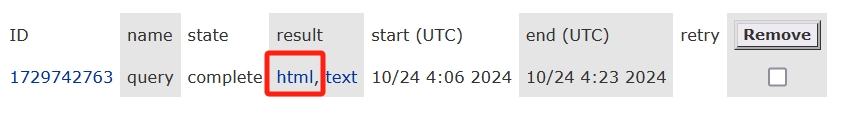

Check out the details:
Check out the details:
Drawing UpSet plot
#!/usr/bin/env Rscript
# -*- coding: utf-8 -*-
# @Author : mengqingyao
# @Time : 20230608
rm(list = ls())
library(tidyfst)
library(readr)
library(readxl)
library(ComplexUpset)
library(tidyverse)
library(gridExtra)
library(eoffice)
# Orthogroups.tsv ---------------------------------------------------------
Orthogroups <- read_tsv("./OrthoFinder/OrthoFinder/Results_May31/Orthogroups/Orthogroups.tsv")
# 去除一个物种
Orthogroups <- Orthogroups %>% select_dt(-`Hartmannula_sinica_185.fna_remove`)
# Orthogroups <- Orthogroups %>% separate_rows("Carteria_cerasiformis_72.fna_remove", sep = ",")
# 分隔变量中的值
Orthogroups <- separate_dt(Orthogroups, "Carteria_cerasiformis_72.fna_remove", c("Carteria_cerasiformis_72_1","Carteria_cerasiformis_72_2","Carteria_cerasiformis_72_3","Carteria_cerasiformis_72_4","Carteria_cerasiformis_72_5"),sep = ",")
Orthogroups <- separate_dt(Orthogroups, "Mesostigma_viride_1.fasta_remove", c("Mesostigma_viride_1_1","Mesostigma_viride_1_2","Mesostigma_viride_1_3","Mesostigma_viride_1_4","Mesostigma_viride_1_5","Mesostigma_viride_1_6","Mesostigma_viride_1_7","Mesostigma_viride_1_8","Mesostigma_viride_1_9","Mesostigma_viride_1_10"),sep = ",")
Orthogroups <- separate_dt(Orthogroups, "Mesostigma_viride_28.fna_remove", c("Mesostigma_viride_28_1","Mesostigma_viride_28_2","Mesostigma_viride_28_3","Mesostigma_viride_28_4","Mesostigma_viride_28_5","Mesostigma_viride_28_6","Mesostigma_viride_28_7","Mesostigma_viride_28_8"),sep = ",")
Orthogroups <- separate_dt(Orthogroups, "Nemacystus_decipiens_23.fna_remove", c("Nemacystus_decipiens_23_1","Nemacystus_decipiens_23_2","Nemacystus_decipiens_23_3","Nemacystus_decipiens_23_4","Nemacystus_decipiens_23_5","Nemacystus_decipiens_23_6","Nemacystus_decipiens_23_7","Nemacystus_decipiens_23_8","Nemacystus_decipiens_23_9","Nemacystus_decipiens_23_10","Nemacystus_decipiens_23_11","Nemacystus_decipiens_23_12","Nemacystus_decipiens_23_13"),sep = ",")
Orthogroups <- separate_dt(Orthogroups, "SAG_1.fna_remove", c("Cryptomonas_gyropyrenoidosa_1_1","Cryptomonas_gyropyrenoidosa_1_2","Cryptomonas_gyropyrenoidosa_1_3","Cryptomonas_gyropyrenoidosa_1_4","Cryptomonas_gyropyrenoidosa_1_5","Cryptomonas_gyropyrenoidosa_1_6","Cryptomonas_gyropyrenoidosa_1_7","Cryptomonas_gyropyrenoidosa_1_8","Cryptomonas_gyropyrenoidosa_1_9","Cryptomonas_gyropyrenoidosa_1_10","Cryptomonas_gyropyrenoidosa_1_11","Cryptomonas_gyropyrenoidosa_1_12",
"Cryptomonas_gyropyrenoidosa_1_13","Cryptomonas_gyropyrenoidosa_1_14","Cryptomonas_gyropyrenoidosa_1_15","Cryptomonas_gyropyrenoidosa_1_16","Cryptomonas_gyropyrenoidosa_1_17","Cryptomonas_gyropyrenoidosa_1_18","Cryptomonas_gyropyrenoidosa_1_19","Cryptomonas_gyropyrenoidosa_1_20","Cryptomonas_gyropyrenoidosa_1_21","Cryptomonas_gyropyrenoidosa_1_22","Cryptomonas_gyropyrenoidosa_1_23","Cryptomonas_gyropyrenoidosa_1_24",
"Cryptomonas_gyropyrenoidosa_1_25","Cryptomonas_gyropyrenoidosa_1_26","Cryptomonas_gyropyrenoidosa_1_27","Cryptomonas_gyropyrenoidosa_1_28","Cryptomonas_gyropyrenoidosa_1_29","Cryptomonas_gyropyrenoidosa_1_30","Cryptomonas_gyropyrenoidosa_1_31","Cryptomonas_gyropyrenoidosa_1_32","Cryptomonas_gyropyrenoidosa_1_33","Cryptomonas_gyropyrenoidosa_1_34","Cryptomonas_gyropyrenoidosa_1_35","Cryptomonas_gyropyrenoidosa_1_36",
"Cryptomonas_gyropyrenoidosa_1_37","Cryptomonas_gyropyrenoidosa_1_38","Cryptomonas_gyropyrenoidosa_1_39","Cryptomonas_gyropyrenoidosa_1_40","Cryptomonas_gyropyrenoidosa_1_41","Cryptomonas_gyropyrenoidosa_1_42","Cryptomonas_gyropyrenoidosa_1_43","Cryptomonas_gyropyrenoidosa_1_44","Cryptomonas_gyropyrenoidosa_1_45","Cryptomonas_gyropyrenoidosa_1_46","Cryptomonas_gyropyrenoidosa_1_47","Cryptomonas_gyropyrenoidosa_1_48",
"Cryptomonas_gyropyrenoidosa_1_49","Cryptomonas_gyropyrenoidosa_1_50","Cryptomonas_gyropyrenoidosa_1_51","Cryptomonas_gyropyrenoidosa_1_52","Cryptomonas_gyropyrenoidosa_1_53"),sep = ",")
Orthogroups <- separate_dt(Orthogroups, "Stentor_roeselii_82.fna_remove", c("Stentor_roeselii_82_1", "Stentor_roeselii_82_2", "Stentor_roeselii_82_3", "Stentor_roeselii_82_4", "Stentor_roeselii_82_5", "Stentor_roeselii_82_6", "Stentor_roeselii_82_7", "Stentor_roeselii_82_8"),sep = ",")
Orthogroups <- separate_dt(Orthogroups, "vtn8_280.fasta_remove", c("E_octocarinatus_VTN8_1", "E_octocarinatus_VTN8_2", "E_octocarinatus_VTN8_3", "E_octocarinatus_VTN8_4", "E_octocarinatus_VTN8_5", "E_octocarinatus_VTN8_6", "E_octocarinatus_VTN8_7", "E_octocarinatus_VTN8_8", "E_octocarinatus_VTN8_9"),sep = ",")
# 选择第一个分列
Orthogroups <- Orthogroups %>% select_dt(Orthogroup,Carteria_cerasiformis_72_1,Mesostigma_viride_1_1,Mesostigma_viride_28_1,Nemacystus_decipiens_23_1,Cryptomonas_gyropyrenoidosa_1_1,E_octocarinatus_VTN8_1,Stentor_roeselii_82_1)
# Orthogroups_UnassignedGenes ---------------------------------------------
Orthogroups_UnassignedGenes <- read_tsv("./OrthoFinder/OrthoFinder/Results_May31/Orthogroups/Orthogroups_UnassignedGenes.tsv")
Orthogroups_UnassignedGenes <- Orthogroups_UnassignedGenes %>% select_dt(-`Hartmannula_sinica_185.fna_remove`)
# 和Orthogroups数据对应
colnames(Orthogroups_UnassignedGenes) <- c("Orthogroup","Carteria_cerasiformis_72_1", "Mesostigma_viride_1_1", "Mesostigma_viride_28_1", "Nemacystus_decipiens_23_1", "Cryptomonas_gyropyrenoidosa_1_1", "Stentor_roeselii_82_1" ,"E_octocarinatus_VTN8_1")
Orthogroups_all <- rbind(Orthogroups,Orthogroups_UnassignedGenes)
#count_dt(Orthogroups_all, Orthogroup)
# all_orthogroup_emapper --------------------------------------------------
# eggnog注释信息
all_7_species_emapper_mod <- read_delim("eggNOG/7_species.emapper_mod.annotations",
delim = "\t", escape_double = FALSE,
trim_ws = TRUE)
# KASS注释信息
Carteria_cerasiformis_72 <- read_delim("KEGG/Carteria_cerasiformis_72.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE, trim_ws = TRUE)
names(Carteria_cerasiformis_72)[2] <- "Carteria_cerasiformis_KO"
names(Carteria_cerasiformis_72)[1] <- "query"
Carteria_cerasiformis_72 <- Carteria_cerasiformis_72 %>% mutate(Carteria_cerasiformis_72 = "1")
Cryptomonas_gyropyrenoidosa_1 <- read_delim("KEGG/Cryptomonas_gyropyrenoidosa_1.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE,trim_ws = TRUE)
names(Cryptomonas_gyropyrenoidosa_1)[2] <- "Cryptomonas_gyropyrenoidosa_KO"
names(Cryptomonas_gyropyrenoidosa_1)[1] <- "query"
Cryptomonas_gyropyrenoidosa_1 <- Cryptomonas_gyropyrenoidosa_1 %>% mutate(Cryptomonas_gyropyrenoidosa_1 = "1")
E_octocarinatus_VTN8 <- read_delim("KEGG/E_octocarinatus_VTN8.txt",
delim = "\t", escape_double = FALSE,
trim_ws = TRUE)
names(E_octocarinatus_VTN8)[2] <- "E_octocarinatus_VTN8_KO"
names(E_octocarinatus_VTN8)[1] <- "query"
E_octocarinatus_VTN8 <- E_octocarinatus_VTN8 %>% mutate(E_octocarinatus_VTN8 = "1")
Mesostigma_viride_1 <- read_delim("KEGG/Mesostigma_viride_1.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE, trim_ws = TRUE)
names(Mesostigma_viride_1)[2] <- "Mesostigma_viride_1_KO"
names(Mesostigma_viride_1)[1] <- "query"
Mesostigma_viride_1 <- Mesostigma_viride_1 %>% mutate(Mesostigma_viride_1 = "1")
Mesostigma_viride_28 <- read_delim("KEGG/Mesostigma_viride_28.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE, trim_ws = TRUE)
names(Mesostigma_viride_28)[2] <- "Mesostigma_viride_28_KO"
names(Mesostigma_viride_28)[1] <- "query"
Mesostigma_viride_28 <- Mesostigma_viride_28 %>% mutate(Mesostigma_viride_28 = "1")
Stentor_roeselii_82 <- read_delim("KEGG/Stentor_roeselii_82.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE, trim_ws = TRUE)
names(Stentor_roeselii_82)[2] <- "Stentor_roeselii_KO"
names(Stentor_roeselii_82)[1] <- "query"
Stentor_roeselii_82 <- Stentor_roeselii_82 %>% mutate(Stentor_roeselii_82 = "1")
Nemacystus_decipiens_23 <- read_delim("KEGG/Nemacystus_decipiens_23.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE, trim_ws = TRUE)
names(Nemacystus_decipiens_23)[2] <- "Nemacystus_decipiens_KO"
names(Nemacystus_decipiens_23)[1] <- "query"
Nemacystus_decipiens_23 <- Nemacystus_decipiens_23 %>% mutate(Nemacystus_decipiens_23 = "1")
# 数据转化 --------------------------------------------------------------------
switch <- merge(all_7_species_emapper_mod, Carteria_cerasiformis_72, by = "query", all=T)
switch <- merge(switch, Cryptomonas_gyropyrenoidosa_1, by = "query", all = T)
switch <- merge(switch, E_octocarinatus_VTN8, by = "query", all = T)
switch <- merge(switch, Mesostigma_viride_1, by = "query", all = T)
switch <- merge(switch, Mesostigma_viride_28, by = "query", all = T)
switch <- merge(switch, Stentor_roeselii_82, by = "query", all = T)
switch <- merge(switch, Nemacystus_decipiens_23, by = "query", all = T)
switch <- switch %>% select_dt(query,eggNOG_OGs,Carteria_cerasiformis_72,Carteria_cerasiformis_KO,
Cryptomonas_gyropyrenoidosa_1,Cryptomonas_gyropyrenoidosa_KO,E_octocarinatus_VTN8,E_octocarinatus_VTN8_KO,
Mesostigma_viride_1,Mesostigma_viride_1_KO,Mesostigma_viride_28,Mesostigma_viride_28_KO,Stentor_roeselii_82,Stentor_roeselii_KO,
Nemacystus_decipiens_23,Nemacystus_decipiens_KO)
# 单个物种分离
Carteria_cerasiformis_72 <- switch %>% filter_dt(Carteria_cerasiformis_72 == "1") %>%
select_dt(query,eggNOG_OGs,Carteria_cerasiformis_KO)
Cryptomonas_gyropyrenoidosa_1 <- switch %>% filter_dt(Cryptomonas_gyropyrenoidosa_1 == "1") %>%
select_dt(query,eggNOG_OGs,Cryptomonas_gyropyrenoidosa_KO)
E_octocarinatus_VTN8 <- switch %>% filter_dt(E_octocarinatus_VTN8 == "1") %>%
select_dt(query,eggNOG_OGs,E_octocarinatus_VTN8_KO)
Mesostigma_viride_1 <- switch %>% filter_dt(Mesostigma_viride_1 == "1") %>%
select_dt(query,eggNOG_OGs,Mesostigma_viride_1_KO)
Mesostigma_viride_28 <- switch %>% filter_dt(Mesostigma_viride_1 == "1") %>%
select_dt(query,eggNOG_OGs,Mesostigma_viride_28_KO)
Nemacystus_decipiens_23 <- switch %>% filter_dt(Nemacystus_decipiens_23 == "1") %>%
select_dt(query,eggNOG_OGs,Nemacystus_decipiens_KO)
Stentor_roeselii_82 <- switch %>% filter_dt(Stentor_roeselii_82 == "1") %>%
select_dt(query,eggNOG_OGs,Stentor_roeselii_KO)
Carteria_cerasiformis_72 <- Orthogroups_all %>% left_join_dt(Carteria_cerasiformis_72,c("Carteria_cerasiformis_72_1" = "query"))
Cryptomonas_gyropyrenoidosa_1 <- Orthogroups_all %>% left_join_dt(Cryptomonas_gyropyrenoidosa_1,c("Cryptomonas_gyropyrenoidosa_1_1" = "query"))
E_octocarinatus_VTN8 <- Orthogroups_all %>% left_join_dt(E_octocarinatus_VTN8,c("E_octocarinatus_VTN8_1" = "query"))
Mesostigma_viride_1 <- Orthogroups_all %>% left_join_dt(Mesostigma_viride_1,c("Mesostigma_viride_1_1" = "query"))
Mesostigma_viride_28 <- Orthogroups_all %>% left_join_dt(Mesostigma_viride_28,c("Mesostigma_viride_28_1" = "query"))
Nemacystus_decipiens_23 <- Orthogroups_all %>% left_join_dt(Nemacystus_decipiens_23,c("Nemacystus_decipiens_23_1" = "query"))
Stentor_roeselii_82 <- Orthogroups_all %>% left_join_dt(Stentor_roeselii_82,c("Stentor_roeselii_82_1" = "query"))
# 按列合并,将不同名的列合成同一列,其他列用NA值填充。
cog_kegg <- rbind(Carteria_cerasiformis_72,Cryptomonas_gyropyrenoidosa_1,E_octocarinatus_VTN8,Mesostigma_viride_1,Mesostigma_viride_28,Nemacystus_decipiens_23,Stentor_roeselii_82,use.names=FALSE )
# 对数据矩阵按重新排序
cog_kegg <- cog_kegg[order(cog_kegg$Orthogroup)]
# 去除Orthogroup中重复的id并保留第一行
cog_kegg <- cog_kegg %>% distinct_dt(Orthogroup,.keep_all = T)
names(cog_kegg)[10] <- "KEGG_id"
cog_kegg <- cog_kegg %>% replace_na_dt(-KEGG_id,-Orthogroup, to = 0)
cog_kegg <- cog_kegg %>%
replace_dt(`Carteria_cerasiformis_72_1`, from = function(x) x != 0 , to = 1 ) %>%
replace_dt(`Mesostigma_viride_1_1`, from = function(x) x != 0 , to = 1 ) %>%
replace_dt(`Mesostigma_viride_28_1`, from = function(x) x != 0 , to = 1 ) %>%
replace_dt(`Nemacystus_decipiens_23_1`, from = function(x) x != 0 , to = 1 ) %>%
replace_dt(`Cryptomonas_gyropyrenoidosa_1_1`, from = function(x) x != 0 , to = 1 ) %>%
replace_dt(`E_octocarinatus_VTN8_1`, from = function(x) x != 0 , to = 1 ) %>%
replace_dt(`Stentor_roeselii_82_1`, from = function(x) x != 0 , to = 1 )
cog_kegg <- cog_kegg %>%
replace_dt(KEGG_id, from = "",to = NA) %>%
replace_na_dt(eggNOG_OGs, KEGG_id, to = 0 ) %>%
replace_dt(KEGG_id, eggNOG_OGs,from = function(x) x != 0 , to = 1)
# 按条件添加新的变量以及观察值(同一变量)
# 以同源基因进行排序
cog_kegg <- cog_kegg[order(cog_kegg[,2])]
# 注释添加分类
cog_kegg[which(cog_kegg$eggNOG_OGs == 1),'class'] <- 'COG'
cog_kegg[which(cog_kegg$KEGG_id == 1 ),'class'] <- 'KEGG'
cog_kegg[which(cog_kegg$eggNOG_OGs == 1 & cog_kegg$KEGG_id == 1),'class'] <- 'COG and KEGG'
cog_kegg <- cog_kegg %>% replace_na_dt(class, to = 1) %>%
replace_dt(class, from = 1, to = "Unknown")
# 选取观测值
cog_kegg <- cog_kegg %>% select_dt(Orthogroup,Carteria_cerasiformis_72_1,Mesostigma_viride_1_1,Mesostigma_viride_28_1,Nemacystus_decipiens_23_1,Cryptomonas_gyropyrenoidosa_1_1,E_octocarinatus_VTN8_1,Stentor_roeselii_82_1,class)
# 消除有NA的一行全部删除,可以通过cols=设置观测值
# filter_dt(Cryptomonas_gyropyrenoidosa_1 != 0 | Carteria_cerasiformis_72 != 0 | Cryptomonas_gyropyrenoidosa_1 != 0 | E_octocarinatus_VTN8 != 0 | Mesostigma_viride_1 != 0 | Mesostigma_viride_28 != 0 | Nemacystus_decipiens_23 != 0 | Stentor_roeselii_82 != 0)
cog_kegg <- cog_kegg %>%
filter_dt(Carteria_cerasiformis_72_1 != 0 | Mesostigma_viride_1_1 != 0 | Mesostigma_viride_28_1 != 0 | Nemacystus_decipiens_23_1 != 0 | Cryptomonas_gyropyrenoidosa_1_1 != 0 | E_octocarinatus_VTN8_1 != 0 | Stentor_roeselii_82_1 != 0)
# cog_kegg <- na.omit(cog_kegg) # 所有的变量都有NA值才能删除
# 字符串类型无法进行计算,转化为字符型。
cog_kegg$`Carteria_cerasiformis_72_1` <- as.numeric(cog_kegg$`Carteria_cerasiformis_72_1`)
cog_kegg$`Mesostigma_viride_1_1` <- as.numeric(cog_kegg$`Mesostigma_viride_1_1`)
cog_kegg$`Mesostigma_viride_28_1` <- as.numeric(cog_kegg$`Mesostigma_viride_28_1`)
cog_kegg$`Nemacystus_decipiens_23_1` <- as.numeric(cog_kegg$`Nemacystus_decipiens_23_1`)
cog_kegg$`Cryptomonas_gyropyrenoidosa_1_1` <- as.numeric(cog_kegg$`Cryptomonas_gyropyrenoidosa_1_1`)
cog_kegg$`E_octocarinatus_VTN8_1` <- as.numeric(cog_kegg$`E_octocarinatus_VTN8_1`)
cog_kegg$`Stentor_roeselii_82_1` <- as.numeric(cog_kegg$`Stentor_roeselii_82_1`)
# cog_kegg %>% count_dt(`Ca. Megaira`)
# cog_kegg %>% count_dt(`Ca. Megaira polyxenophila`)
# cog_kegg %>% count_dt(`Ca. Megaira Meg NIES296`)
# upset -------------------------------------------------------------------
# 增加颜色映射
CC <- c(`COG` = '#3C5488B2', `COG and KEGG` = '#4DBBD5B2', `KEGG` = '#FF3333', `Unknown` = '#DDDDDD')
# 设置物种分类
special <- colnames(cog_kegg)[2:8]
# 画图
p1 <- upset(cog_kegg,
special,
width = .1,
wrap = T,
#min_size = 12,
# 按照交集类型和交集大小排列
sort_intersections_by=c('degree', 'cardinality'),
base_annotations = list(
"intersection size" = intersection_size(
counts = T,
mapping = aes(fill=class)
)
+theme(axis.line.y = element_line(colour = "black",size=0.5),axis.ticks.y=element_line(colour = "black",size=0.5))+
scale_y_continuous(expand = c(0,0))+
scale_fill_manual(values = CC)
),
set_sizes=(
upset_set_size()+ geom_text(aes(label=..count..), hjust=1, stat='count')
+ expand_limits(y=2300)
+ theme(axis.text.x=element_text(angle=90))
))
ggsave(filename = "test.pdf", p1, width = 60, height = 30, units = "cm")RStudio is recommended for running this script.
- There is a huge error and this can lead to a lot of annotation information loss.
- Fixed bugs and updated code. Click
I wrote this script a long time ago, and there is a lot of redundant code, but this is the original processing logic.
It's better to understand.
Result


Beautification
- I would suggest saving the image as a pdf.
- Evolutionary landscaping using Adobe Illustrator.
- Save images as needed.
Quote
Email me with more questions! 584338215@qq.com
