ggtree: KEGG Pathway Heatmap!
Introduction
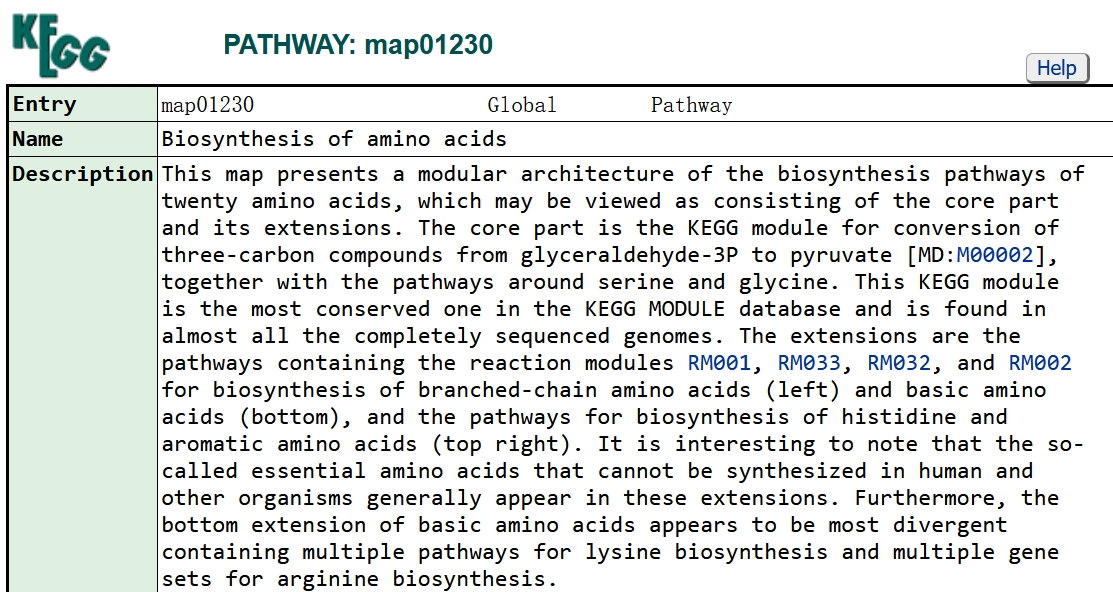
KEGG pathway and module correspondence information. Click
Data preparation
- tools_RStudio_result_all
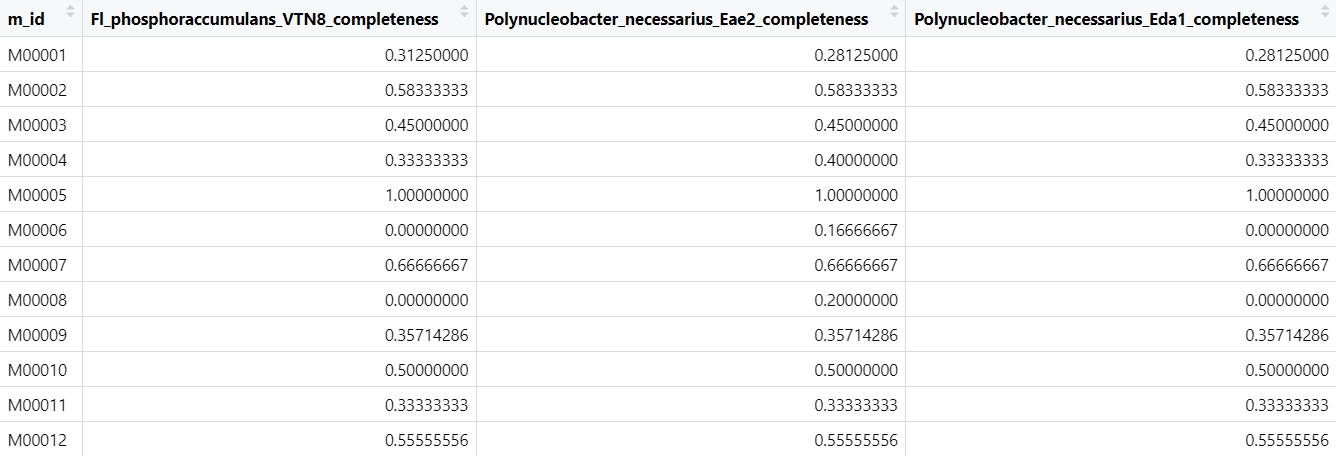
- Maps_Moules_2023_11_16

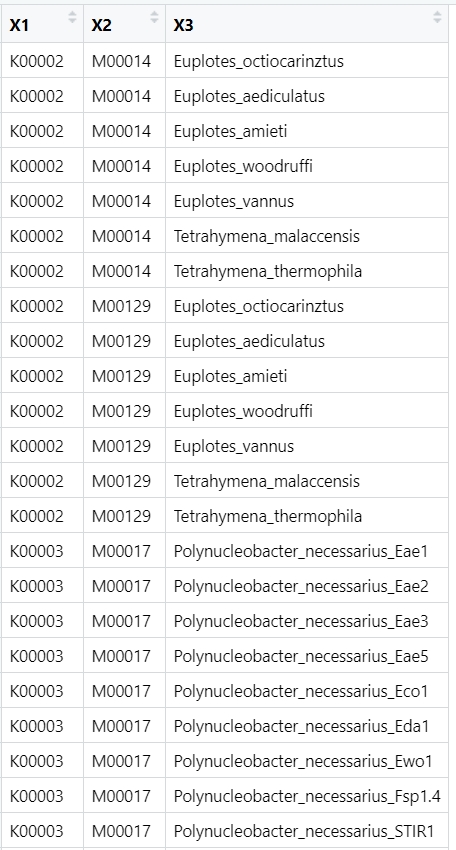
- tools_RStudio_映射模块上的K号
R script for tree + heatmap
#!/usr/bin/env Rscript
# -*- coding: utf-8 -*-
# @Author : mengqingyao
# @Time : 20231116
library(readr)
library(tidyfst)
tools_RStudio_result_all <- read_delim("tools_RStudio_result_all.txt",
delim = "\t", escape_double = FALSE,
trim_ws = TRUE)
Maps_Moules_2023_11_16 <- read_delim("F:/database/KEGG/KEGG_General_Documentation_module/Maps_Moules_2023_11_16.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE, trim_ws = TRUE)
tools_RStudio_映射模块上的K号 <- read_delim("tools_RStudio_映射模块上的K号.txt",
delim = "\t", escape_double = FALSE,
col_names = FALSE, trim_ws = TRUE)
map01230 <- Maps_Moules_2023_11_16 %>% filter_dt(X2 == "map01230")
map01230 <- map01230 %>% rename_dt(m_id = X1)
map01230_1 <- map01230 %>% inner_join_dt(tools_RStudio_result_all,by = "m_id")
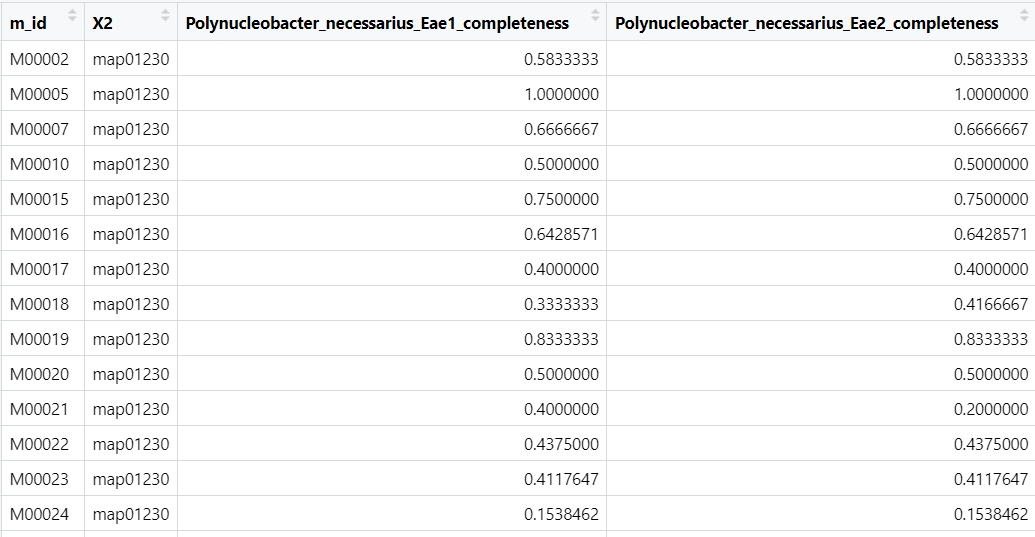
write.table(map01230_1,file = "map01230.txt",sep = "\t",quote = F,row.names = F)- map01230.txt
#!/usr/bin/env Rscript
# -*- coding: utf-8 -*-
# @Author : mengqingyao
# @Time : 20231118
library(ggtree)
library(ggtreeExtra)
library(tidyfst)
map01230 <- read_delim("map01230.txt", delim = "\t",
escape_double = FALSE, trim_ws = TRUE)
map01230 <- map01230 %>% select(-X2)
head_data <- map01230 %>% longer_dt(name = "species",value = "complet")
a <- map01230 %>% column_to_rownames(var="m_name")
tree <- map01230 %>% column_to_rownames(var="m_name") %>%
# 该函数使用指定的距离度量计算数据矩阵行间的距离,计算并返回距离矩阵。
dist() %>%
# Tree Estimation Based on an Improved Version of the NJ Algorithm
ape::bionj()
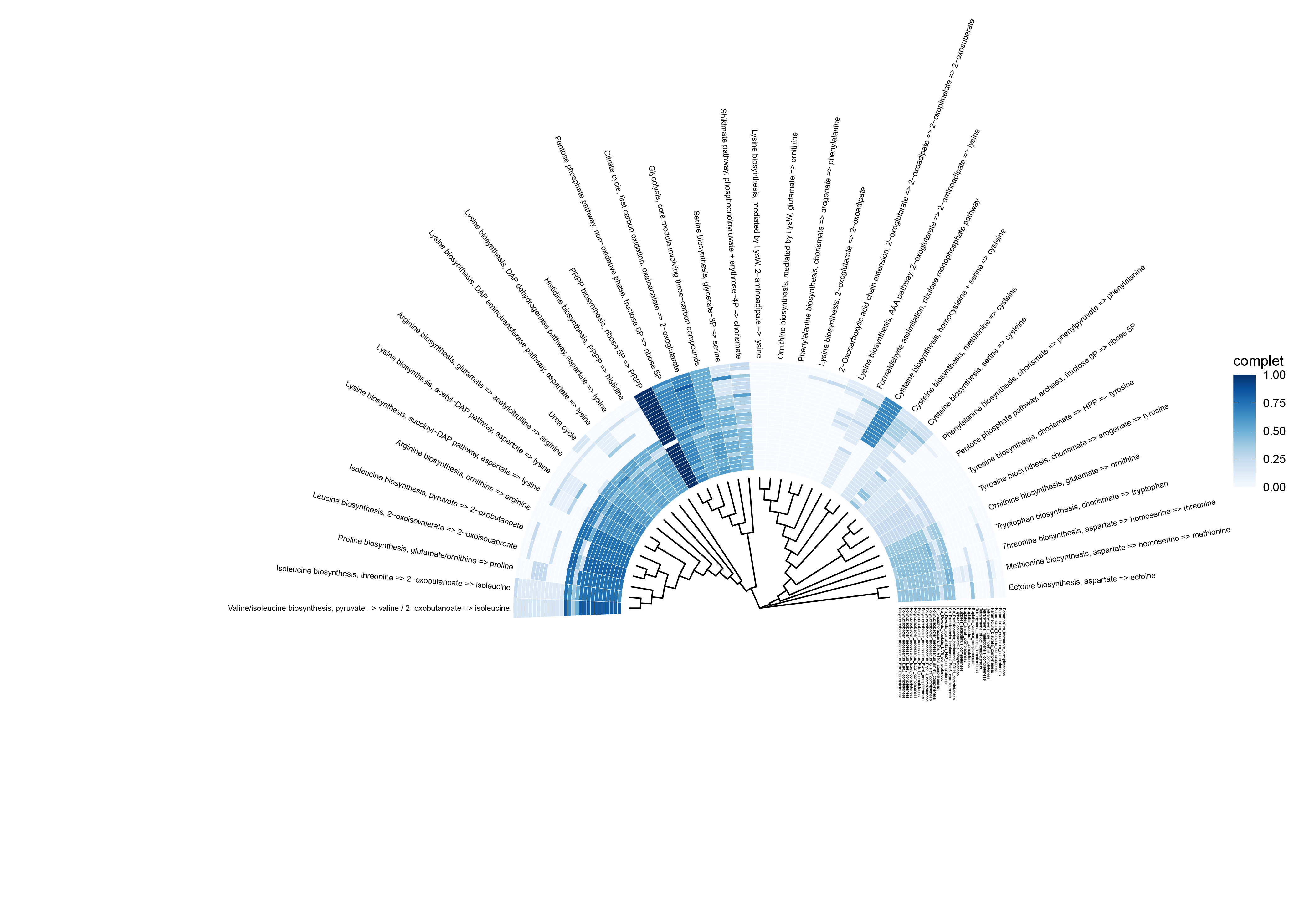
ggtree(tree,branch.length = "none", layout = "circular",
linetype = 1,size = 0.5, ladderize = T)+
layout_fan(angle =180)+ # 调整开口
geom_tiplab(offset=11,show.legend=FALSE,size=2,
color = "black",starstroke = 0)+
geom_fruit(data=head_data,
geom=geom_tile,
mapping=aes(y=m_name,x=species,fill=complet),
pwidth=0.8,
offset=0.04,
color = "white",
axis.params=list(axis="x",text.angle=-90,text.size=1,hjust=0))+
scale_fill_gradientn(colours = RColorBrewer::brewer.pal(100,"Blues")) +
theme(plot.margin=margin(8,-6,0,0,"cm"),# 上下,左右,远近
legend.position = c(1.3,0.8),
legend.background = element_rect(fill = NA),
)Rsudio is recommended for running this script.
I wrote this script a long time ago, and there is a lot of redundant code, but this is the original processing logic.
It's better to understand.
Result
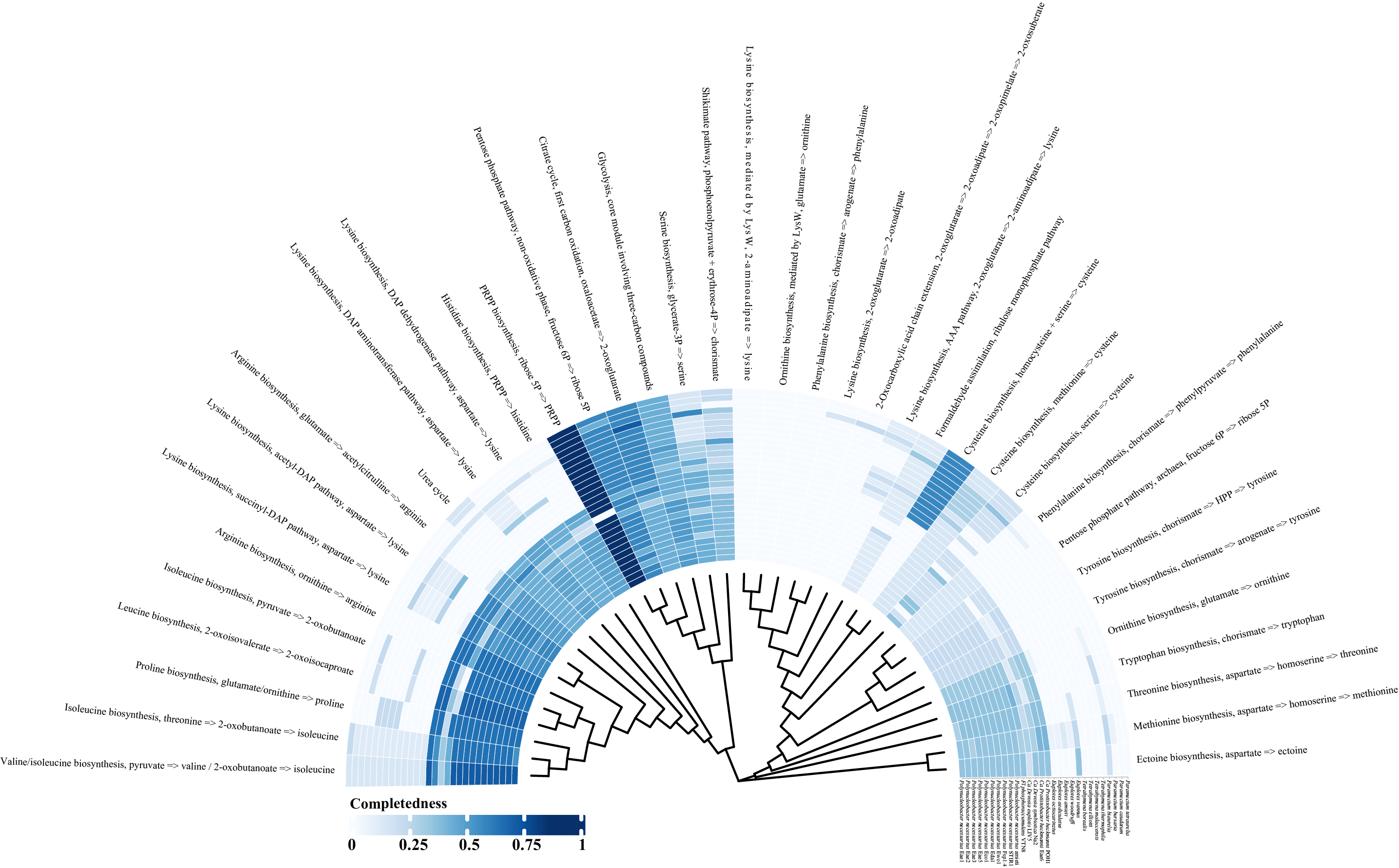
Beautification
- I would suggest saving the image as a pdf.
- Evolutionary landscaping using Adobe Illustrator.
- Save images as needed.
Quote
Email me with more questions! 584338215@qq.com
