GenBank database download tool!
Introduction
Download using the wget utility, which supports broken connections and multiple connection attempts, the key to success is the speed and stability of the network.
This script supports downloadable sequence formats and Blast-specific formats for a total of three databases: nucleic acid, protein and swissprot.
Code example
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# @Author : mengqingyao
# @Email : 15877464851@163.com
# @Time : 2024/11/20
import requests
import subprocess
import os
import glob
import argparse
import sys
from tqdm import tqdm
import time
from datetime import datetime
# 定义两个不同的数据库URL
NR_METADATA_URL = "https://ftp.ncbi.nlm.nih.gov/blast/db/nr-prot-metadata.json"
NT_METADATA_URL = "https://ftp.ncbi.nlm.nih.gov/blast/db/nt-nucl-metadata.json"
ADDITIONAL_URLS = {
"nr_fasta": "https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz",
"nt_fasta": "https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nt.gz",
"swissprot": "https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/swissprot.gz",
}
DATABASE_URLS = {
"nr": NR_METADATA_URL,
"nt": NT_METADATA_URL,
}
def execute_command(command, check=True):
"""执行命令并处理异常"""
try:
process = subprocess.run(command, stdout=subprocess.PIPE, stderr=subprocess.STDOUT, universal_newlines=True, check=check)
return process.stdout
except subprocess.CalledProcessError as e:
print(f"命令执行失败: {e}")
return None
def download_database(metadata_url, output_dir):
response = requests.get(metadata_url)
if response.status_code != 200:
print(f"请求失败,状态码:{response.status_code}")
return False
data = response.json()
files = data.get("files", [])
os.makedirs(output_dir, exist_ok=True)
file_links_path = os.path.join(output_dir, "file_links.txt")
with open(file_links_path, "w") as f:
f.write("\n".join(files) + "\n")
print(f"{metadata_url} 的文件链接已成功写入 {file_links_path}。")
file_links_path = os.path.join(output_dir, "file_links.txt")
# 将文件链接的前缀替换为 https
files_with_https = [file.replace("ftp://", "https://") for file in files]
with open(file_links_path, "w") as f:
f.write("\n".join(files_with_https) + "\n")
print(f"{metadata_url} 的文件链接已成功写入 {file_links_path}。")
print(f"开始下载 {metadata_url} 的文件...")
# 假设 args.output_dir 是你传入的输出目录
output = execute_command(["wget", "-c", "--retry-connrefused", "--tries", "0", "-t", "0", "-P", output_dir, "-i", file_links_path])
if output is not None:
print("下载输出:")
print(output) # 输出下载日志
total_files = len(files)
downloaded_files = output.count("100%")
if downloaded_files == total_files:
print("所有文件下载完成。")
return True
else:
print(f"下载完成的文件数量与预期不符:{downloaded_files}/{total_files}。")
return False
return False
def download_additional_file(file_url, output_dir):
os.makedirs(output_dir, exist_ok=True)
print(f"开始下载 {file_url}...")
output = execute_command(["wget", "-c", "--retry-connrefused", "--tries", "0", file_url,"-P", output_dir])
if output and "100%" in output:
print(f"{file_url} 下载完成。")
return True
else:
print(f"{file_url} 下载过程中出现错误。")
print("下载输出:")
print(output)
return False
def extract_files(output_dir):
downloaded_files = glob.glob(os.path.join(output_dir, "*.tar.gz"))
if not downloaded_files:
print("没有找到需要解压的文件。")
return
print("开始解压下载的文件...")
for tar_file in downloaded_files:
execute_command(["tar", "-xzvf", tar_file])
print(f"{tar_file} 解压完成。")
os.remove(tar_file)
print(f"已删除文件: {tar_file}")
def main():
current_time = datetime.now().strftime("%Y年%m月%d日 %H:%M:%S")
print(f"\n\t\t\t{current_time}\n")
parser = argparse.ArgumentParser(description="Download and extract NCBI BLAST databases.",
epilog="更新数据库信息,如果需要构建diamond数据库,请下载fasta序列文件。")
parser.add_argument("--blast_db", choices=DATABASE_URLS.keys(), help="选择要下载的数据库,支持:'nr' 或 'nt'。")
parser.add_argument("--fasta", choices=ADDITIONAL_URLS.keys(), help="选择要下载的额外文件,支持:'nr_fasta', 'nt_fasta', 'swissprot'。")
parser.add_argument("--output_dir", type=str, required=True, help="输出数据库文件的目录.")
args = parser.parse_args()
start_time = time.time() # 记录开始时间
if args.blast_db:
print(f"开始下载数据库: {args.blast_db}")
download_success = download_database(DATABASE_URLS[args.blast_db], args.output_dir)
if download_success:
extract_files(args.output_dir)
if args.fasta:
print(f"开始下载额外的文件: {args.fasta}")
download_success = download_additional_file(ADDITIONAL_URLS[args.fasta], args.output_dir)
if download_success:
extract_files(args.output_dir)
elapsed_time = time.time() - start_time # 计算总共的运行时间
hours, remainder = divmod(elapsed_time, 3600)
minutes, seconds = divmod(remainder, 60)
print(f"总运行时间: {int(hours)}h {int(minutes)}m {int(seconds)}s") # 输出运行时间
if __name__ == "__main__":
main()Paste copy the above script into a file and it will work, for example:
NCBI_database_down.pyThe type of information that can be accessed.
python ./NCBI_database_down.py -h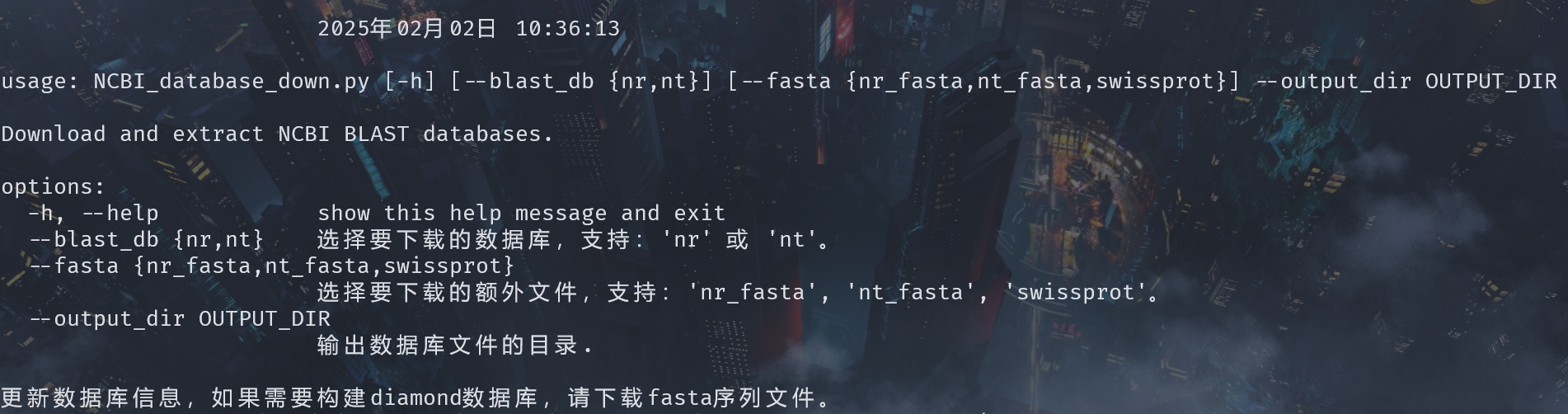
- blast_db: Get information about nucleic acid and protein databases of blast.
- fasta: Get information about nucleic acid and protein databases of sequences.
- output_dir: Customize the output directory.
Results
Download protein databases of blast.
python ./NCBI_database_down.py --blast_db nr -o nr_blast_db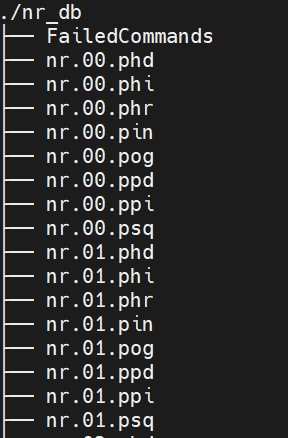
Download nucleic acid databases of sequences.
python ./NCBI_database_down.py --fasta nt_fasta -o nt_dbEmail me with more questions! 584338215@qq.com
