KEGG annotation information collection and update!
Introduction
Since the kegg database is updated quickly, we need to keep its information up to date.
Just do it.
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# @Author : mengqingyao
# @Email : 15877464851@163.com
# @Time : 2024/11/09
import requests
import time
import datetime
from concurrent.futures import ThreadPoolExecutor, as_completed
import json
import sys
import re
import argparse
current_date = datetime.datetime.now().strftime("%Y-%m-%d %H:%M:%S")
def print_colored(text, color):
color_codes = {
'purple': '\033[95m',
'green': '\033[92m',
'red': '\033[91m',
'reset': '\033[0m'
}
print(f"{color_codes.get(color, '')}{text}{color_codes['reset']}")
def fetch_json(url):
try:
response = requests.get(url)
response.raise_for_status()
return response.json()
except requests.RequestException as e:
print(f"请求失败,错误信息:{e}")
except json.JSONDecodeError:
print("JSON解析失败")
return None
def write_output(file_path, header, lines):
try:
with open(file_path, 'w') as outFile:
outFile.write(header + '\n')
outFile.writelines(lines)
except IOError as e:
print(f"写入文件失败,错误信息:{e}")
def extract_module_name_and_path(module_info, ml):
module_name = ml[1].split('[')[0] if re.search(r'\[PATH:', module_info) else ml[1]
relatePath = re.findall(r'\[PATH:.*?]', module_info)[0].split(":")[1].strip(']') if module_name != ml[1] else 'NA'
return module_name, relatePath
def fetch_and_process_kegg_module(output_file, output_file_flag=True):
url = "https://www.kegg.jp/kegg-bin/download_htext?htext=ko00002&format=json&filedir="
module = fetch_json(url)
if not module:
if output_file_flag:
write_output(output_file, 'moduleID\tdescription\tpathway\tlevel0\tlevel1\tlevel2', [])
return []
module_info = ['moduleID\tdescription\tpathway\tlevel0\tlevel1\tlevel2']
for level0 in module['children']:
for level1 in level0['children']:
for level2 in level1['children']:
for level3 in level2['children']:
module_info_entry = level3['name'].strip()
ml = module_info_entry.split(' ')
moduleID = ml[0]
module_name, relatePath = extract_module_name_and_path(module_info_entry, ml)
line = '%s\t%s\t%s\t%s\t%s\t%s\n' % (
moduleID,
module_name,
relatePath,
level0["name"],
level1["name"],
level2["name"]
)
module_info.append(line)
# 将构建好的行写入文件
if output_file_flag:
write_output(output_file, module_info[0], module_info[1:])
return module_info
def fetch_and_process_kegg_pathway(output_file, output_file_flag=True):
url = "https://www.kegg.jp/kegg-bin/download_htext?htext=br08901&format=json&filedir="
pathway = fetch_json(url)
if not pathway:
if output_file_flag:
write_output(output_file, 'mapID\tdescription\tlevel1\tlevel2', [])
return []
pathway_info = ['mapID\tdescription\tlevel1\tlevel2']
if pathway:
for level0 in pathway.get('children', []):
for level1 in level0.get('children', []):
for level2 in level1.get('children', []):
level2_name = level2.get('name', '').strip()
ll = level2_name.split(' ')
if len(ll) >= 2:
line = 'map%s\t%s\t%s\t%s\n' % (
ll[0],
ll[1],
level0.get("name", "NA"),
level1.get("name", "NA")
)
pathway_info.append(line)
else:
print(f"警告:未能正确解析level2名称:{level2_name}")
# 将构建好的行写入文件
if output_file_flag:
write_output(output_file, pathway_info[0], pathway_info[1:])
return pathway_info
def fetch_and_process_kegg_compounds(output_file):
url = "https://www.genome.jp/kegg-bin/download_htext?htext=br08001&format=json&filedir="
compounds = fetch_json(url)
lines = ['CompoundID\tdescription\tlevel1\tlevel2\tlevel3']
if compounds:
for level0 in compounds.get('children', []):
for level1 in level0.get('children', []):
for level2 in level1.get('children', []):
for level3 in level2.get('children', []):
cl = level3.get('name', '').strip().split(' ')
if len(cl) >= 2:
line = '%s\t%s\t%s\t%s\t%s\n' % (
cl[0],
cl[1],
level0.get("name", "NA"),
level1.get("name", "NA"),
level2.get("name", "NA")
)
lines.append(line)
else:
print(f"警告:未能正确解析level3名称:{level3.get('name', '')}")
write_output(output_file, lines[0], lines[1:])
def fetch_data_from_kegg(url, pathway_id):
max_retries = 3
retry_count = 0
while retry_count < max_retries:
try:
response = requests.get(url)
response.raise_for_status()
values = []
for content_line in response.text.splitlines():
if content_line.strip():
al = content_line.split('\t')
if len(al) > 1:
value = al[1].strip().split(':')[-1] if al[1].strip().startswith("ko:") or al[1].strip().startswith("cpd:") else al[1].strip()
values.append(value)
return values
except requests.RequestException as e:
retry_count += 1
print(f"获取数据失败,模块ID: {pathway_id}, 错误信息:{e},正在尝试重新连接,当前尝试次数:{retry_count}/{max_retries}")
if retry_count >= max_retries:
return []
return []
def process_kegg_links(module_info, output_file, data_type):
if not module_info or len(module_info) < 2:
print("警告:输入的模块信息为空,无法处理。")
return
module_ko = {}
module_comp = {}
total_modules = len(module_info) - 1
processed_modules = 0
for line in module_info[1:]:
fields = line.split('\t')
module_id = fields[0]
ko_url = f"http://rest.kegg.jp/link/ko/{module_id}"
module_ko[module_id] = fetch_data_from_kegg(ko_url, module_id)
time.sleep(1)
comp_url = f"http://rest.kegg.jp/link/compound/{module_id}"
module_comp[module_id] = fetch_data_from_kegg(comp_url, module_id)
time.sleep(1)
processed_modules += 1
progress = (processed_modules / total_modules) * 100
print(f"{data_type}处理进度: {progress:.2f}% ({processed_modules}/{total_modules})")
with open(output_file, 'w') as outFile:
if data_type == "模块":
header = module_info[0].strip() + '\tkoList\tcompoundsList\n'
else:
header = 'mapID\tdescription\tlevel1\tlevel2\tkoList\tcompoundsList\n'
outFile.write(header)
for line in module_info[1:]:
if line.strip():
bl = line.strip().split('\t')
kolist = module_ko.get(bl[0], [])
complist = module_comp.get(bl[0], [])
outFile.write('\t'.join(bl) + '\t' + ','.join(kolist) + '\t' + ','.join(complist) + '\n')
def process_kegg_links_module(module_info, output_file):
process_kegg_links(module_info, output_file, data_type="模块")
def process_kegg_links_pathway(pathway_info, output_file):
process_kegg_links(pathway_info, output_file, data_type="通路")
if __name__ == "__main__":
print_colored("\n 由于kegg数据库更新的速度较快,因此我们需要不断更新其信息。\n", 'purple')
print_colored(" 此脚本用于获取并输出KEGG模块、通路和化合物信息。\n", 'purple')
print_colored(" >>> 注意:五种参数独立使用! <<<", 'red')
print_colored(" >>> 注意:五种参数必须选择一种! <<<\n", 'red')
print_colored(" [link 模式]\n", 'purple')
print_colored(" >>> 注意:由于KEGG网站的访问频率限制,link模式下运行速度可能受到一定影响。 <<<\n", 'red')
print_colored(f" 当前日期: {current_date}", 'green')
parser = argparse.ArgumentParser(description=print_colored('\t\t [感谢使用本脚本]\n','green'),
epilog=print_colored('\t更详细的信息请访问: https://mengqy2022.github.io/gene%20annotation/database/kegg-infomation/\n','green'))
parser.add_argument('-o', '--output', required=True, help='指定输出文件的名称')
parser.add_argument('--modules', action='store_true', help='获取模块信息')
parser.add_argument('--pathways', action='store_true', help='获取通路信息')
parser.add_argument('--compounds', action='store_true', help='获取化合物信息')
parser.add_argument('--module-links', action='store_true', help='获取模块与化合物、KO的链接信息')
parser.add_argument('--pathway-links', action='store_true', help='获取路径与化合物、KO的链接信息')
args = parser.parse_args()
if not (args.modules or args.pathways or args.compounds or args.module_links or args.pathway_links):
print_colored("请至少选择一个数据类型:--modules, --pathways, --compounds, --module-links, --pathway-links", 'red')
sys.exit(1)
selected_count = sum([args.modules, args.pathways, args.compounds, args.module_links, args.pathway_links])
if selected_count > 1:
print_colored("错误:只能选择一个数据类型,您选择了多个!\n", 'red')
sys.exit(1)
try:
if args.modules:
module_info = fetch_and_process_kegg_module(args.output)
if args.module_links:
module_info = fetch_and_process_kegg_module(args.output, output_file_flag=False)
if not module_info or len(module_info) < 2:
print("警告:获取的模块信息为空或数据不完整,无法处理模块链接。")
else:
process_kegg_links_module(module_info, args.output)
if args.pathways:
pathway_info = fetch_and_process_kegg_pathway(args.output)
if args.pathway_links:
pathway_info = fetch_and_process_kegg_pathway(args.output, output_file_flag=False)
process_kegg_links_pathway(pathway_info, args.output)
if args.compounds:
fetch_and_process_kegg_compounds(args.output)
except Exception as e:
print(f"处理过程中发生错误:{e}")
sys.exit(1)Paste copy the above script into a file and it will work, for example:
python ./kegg_info_inte_one_upgrade.py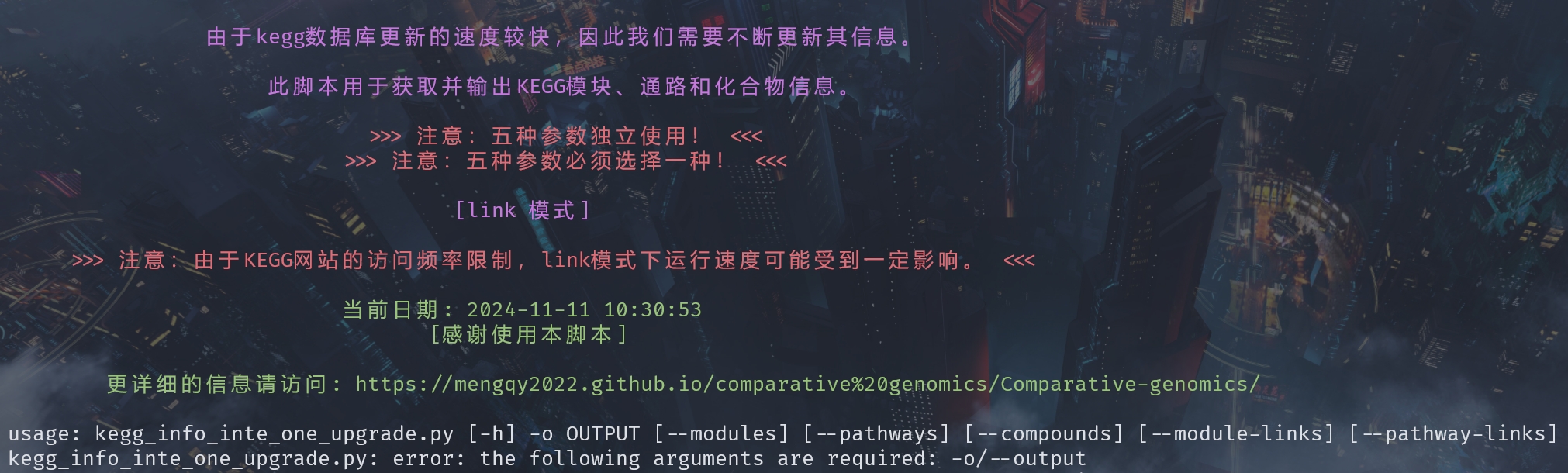
The type of information that can be accessed.
python ./kegg_info_inte_one_upgrade.py -h
- Modules: Get information about the KEGG module classification.
- Pathways: Get the information of KEGG pathways classification.
- Compounds: Get the information of KEGG compounds classification.
- Module Links: Get the links between KEGG modules and compounds and KOs.
- Pathway Links: Get the links between KEGG pathways and compounds and KOs.
Results
python ./kegg_info_inte_one_upgrade.py --module-links -o test_moudle_link.txt
python ./kegg_info_inte_one_upgrade.py --pathway-links -o test_pathway_link.txt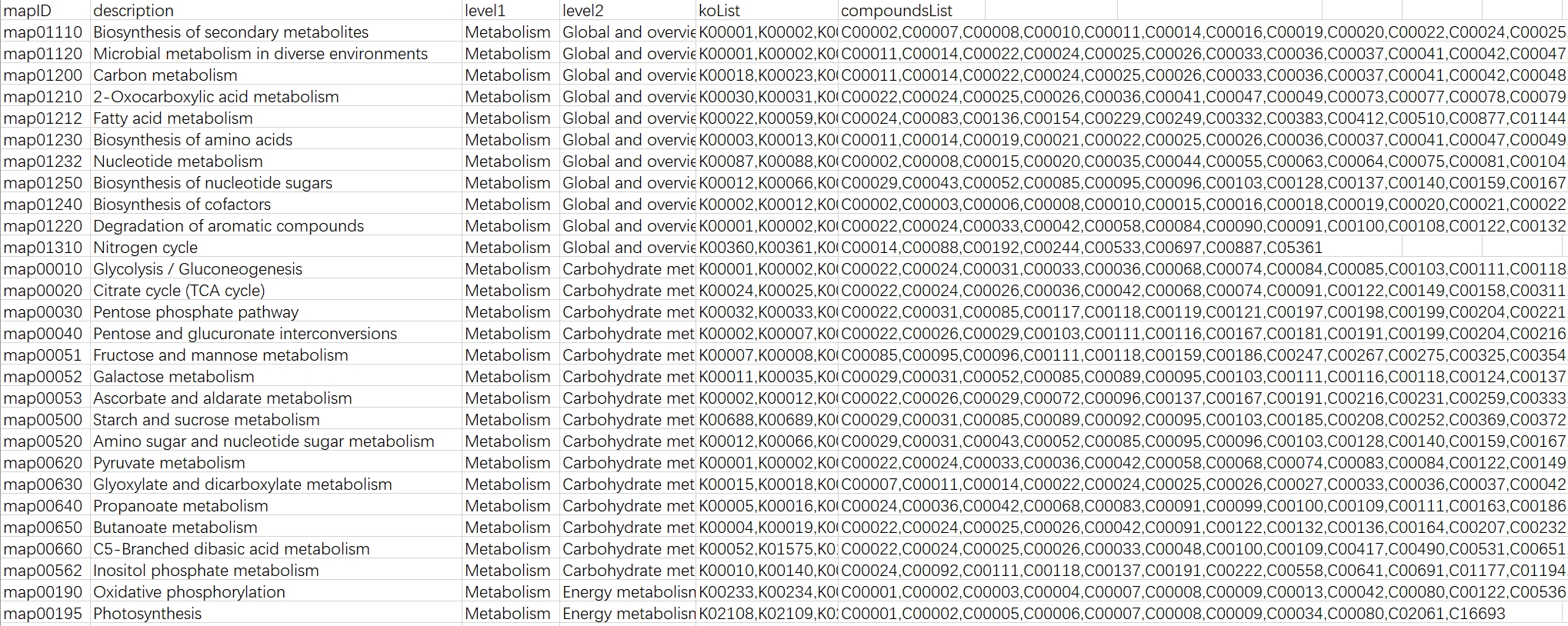
python ./kegg_info_inte_one_upgrade.py --modules -o modules.txt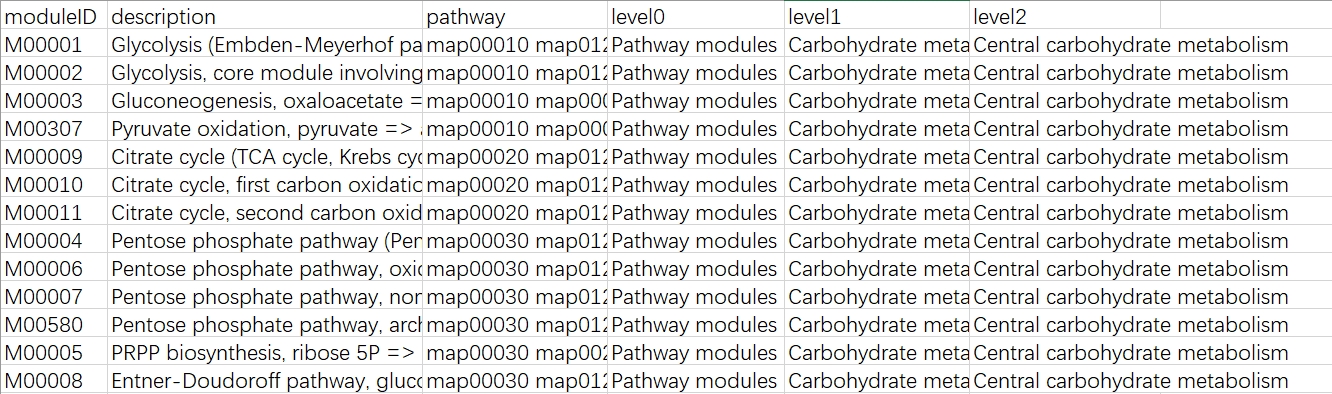
This information can be used for further analysis and visualization. barplot heatmap
python ./kegg_info_inte_one_upgrade.py --pathways -o pathways.txt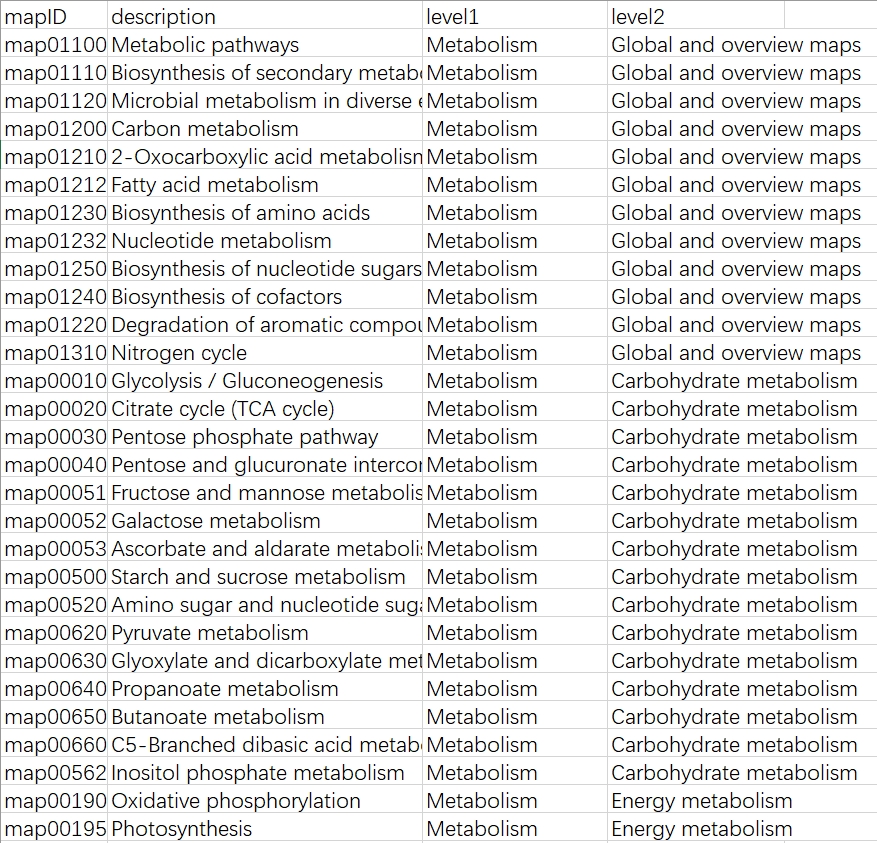
python ./kegg_info_inte_one_upgrade.py --compounds -o compounds.txt
Email me with more questions! 584338215@qq.com
