Eggnog-mapper Gene Function Annotation!
Introduction
Gene function annotation, in simple terms, is the comparison of protein sequences extracted from the genome based on existing databases to obtain the corresponding information.
- eggNOG
- The whole genome protein sequences of 5090 organisms (477 eukaryotes, 4445 representative bacteria and 168 archaea) and 2502 viruses were collected in the eggNOG public database version V5.0.
- These species were grouped into 379 categories (taxonomic levels), with the number of each category indicated by the NCBI classification number.
- Homologous gene classification of the whole genome protein sequences of species in each category yielded a total of 4.4M homologous gene classes. eggNOG database is an extension of NCBI’s COG database, which collects a more comprehensive set of species and a larger amount of protein sequence data.
- The eggNOG database provides a comprehensive set of gene function annotations, including the functional categories of each gene (COG), the gene ontology (GO) terms, and the KEGG pathway information.
Previously we obtained protein coding sequences through the bacterial genome Click, and now we want to construct a single-copy genome phylogenetic tree to analyze the evolutionary relationships among bacteria.
Go to the next step
Obtain the protein coding sequences
- First, we can download the bacterial protein coding sequences from any public databases.
- Secondly, genome annotation was performed to obtain protein coding sequences.
eggNOG-mapper Gene function annotation
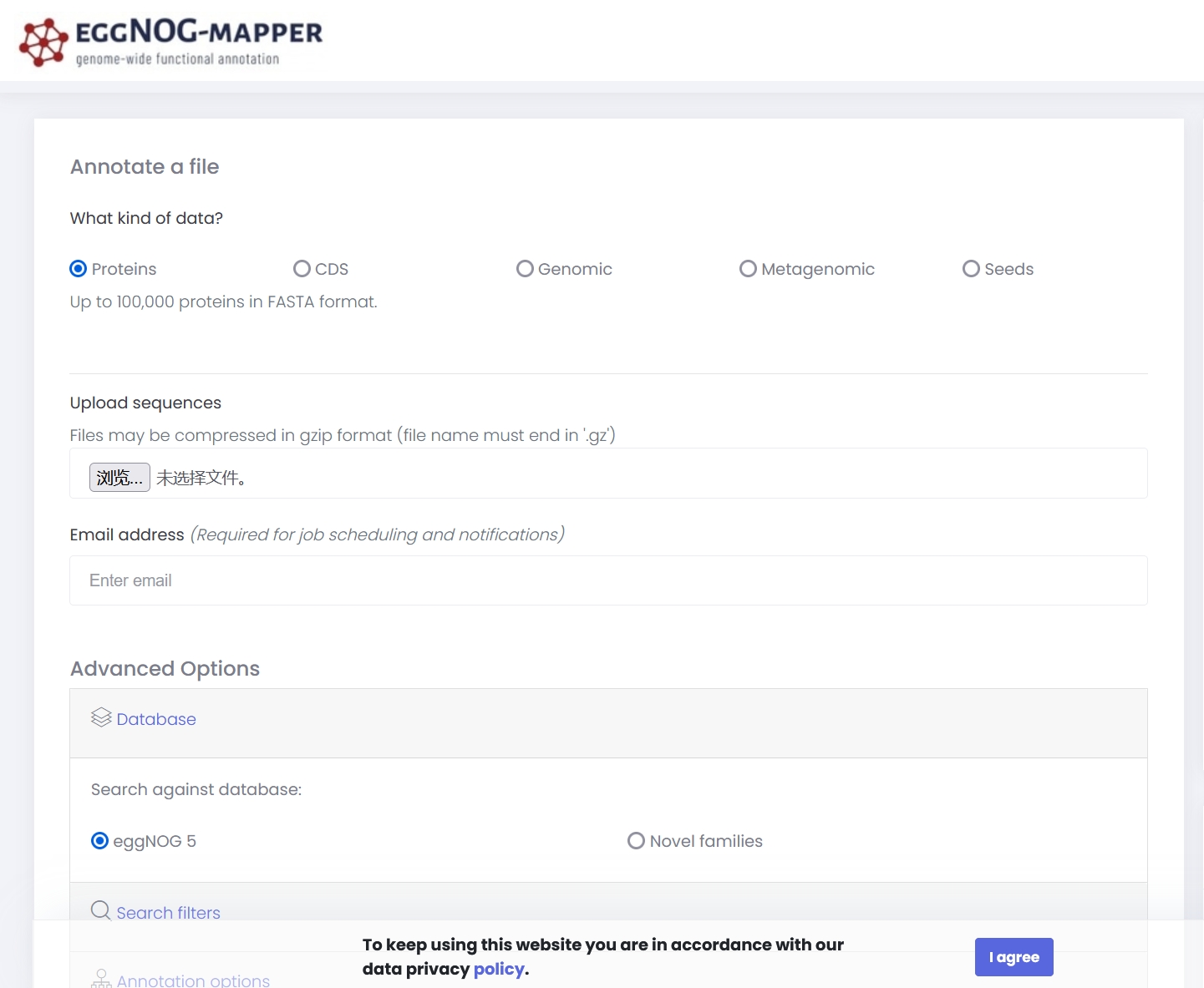
Online Annotation
Check if eggnog-mapper is installed.
- Select sequence type
- Upload the protein coding sequence file
- Enter your e-mail address
Search filtersandAnnotation optionscan be adjusted as needed.- Click
Submitto start the annotation process.
Wait for the completion of the job.
Download the result file for your email.
Local Annotation
- Need download the eggNOG-mapper software.
- Download the eggNOG database.
- Run the eggNOG-mapper software.
- Check if eggnog-mapper is installed.
emapper.py -v
- Download the eggNOG database.
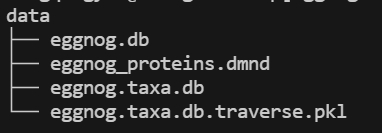
You need set the database path in the emapper.py configuration file.
- The software requires diamond support.
diamond help
Diamond database
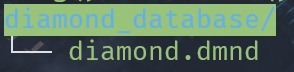
- Run the eggNOG-mapper software.
nohup python3 emapper.py -m diamond -i input.fa -o output.txt --tax_scope all --cpu 0 &results
Quote
Email me with more questions! 584338215@qq.com
