KEGG Gene Function Annotation Submit!
Introduction
Gene function annotation, in simple terms, is the comparison of protein sequences extracted from the genome based on existing databases to obtain the corresponding information.
KEGG is a resource of utility databases for understanding high-level functions and biological systems (e.g., cells, organisms, and ecosystems) from molecular-level information, especially large molecular datasets generated by genome sequencing and other high-throughput experimental techniques, established by the Kanehisa Laboratory at the Center for Bioinformatics, Kyoto University, Japan, in 1995.
KAAS (KEGG Automatic Annotation Server) provides functional annotation of genes by BLAST or GHOST comparisons against the manually curated KEGG GENES database. The result contains KO (KEGG Orthology) assignments and automatically generated KEGG pathways.
Previously we obtained protein coding sequences through the bacterial genome Click, and now we want to construct a single-copy genome phylogenetic tree to analyze the evolutionary relationships among bacteria.
Use
Obtain the protein coding sequences
- First, we can download the bacterial protein coding sequences from any public databases.
- Secondly, genome annotation was performed to obtain protein coding sequences.
Click on the function we need.
Complete or Draft Genome
Let’s now take Complete or Draft Genome as an example and click on KAAS job request (BBH method)
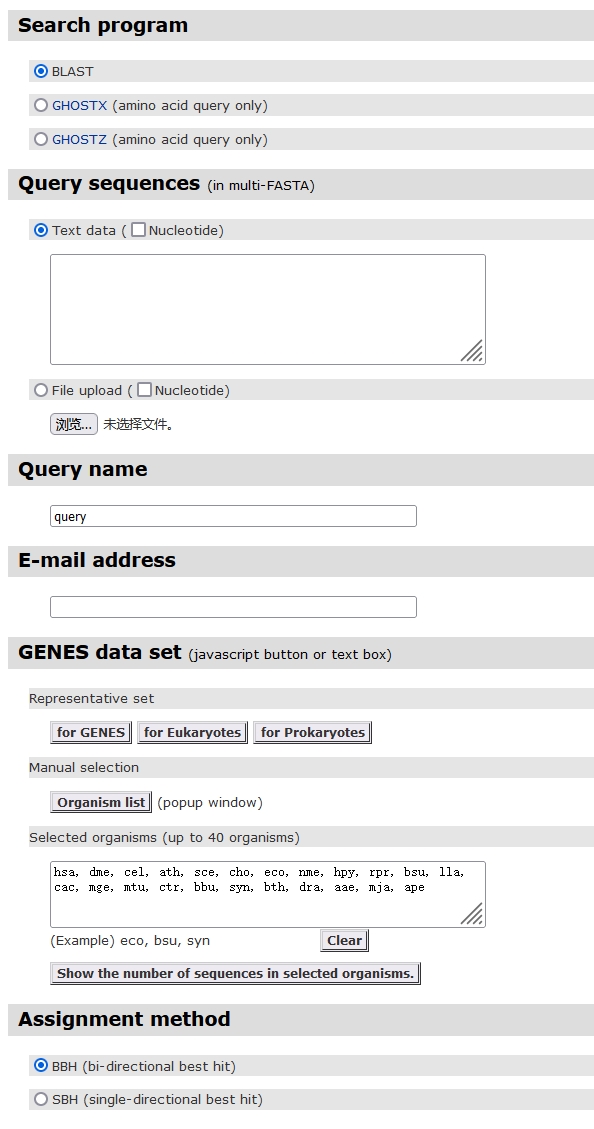
If you don’t know these parameters particularly well, simply fill in some of them.
- Query sequences:
- Select the file or paste the sequence into it.
- Query name:
- The name of the query sequence.
- E-mail address:
- Your email address.
Submission of protein sequences is required.
Now you can complete the job request.
Check your mail
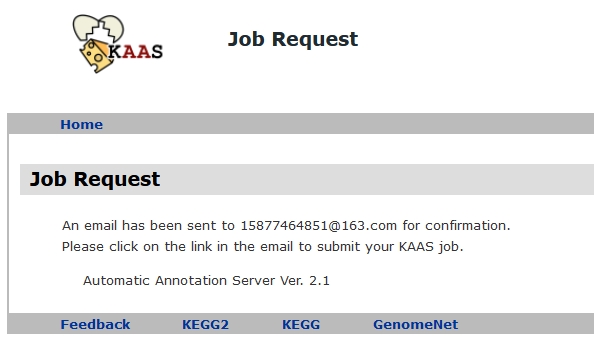
Now log in to your email to find new messages
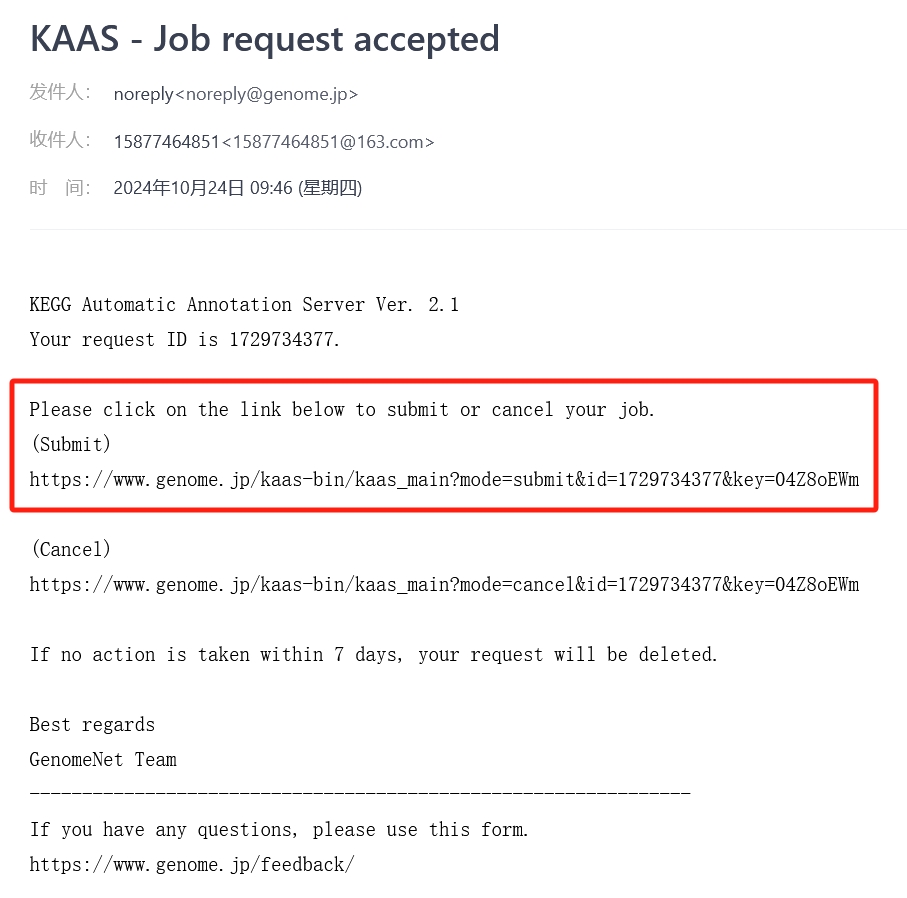
Access to the “Submit” site
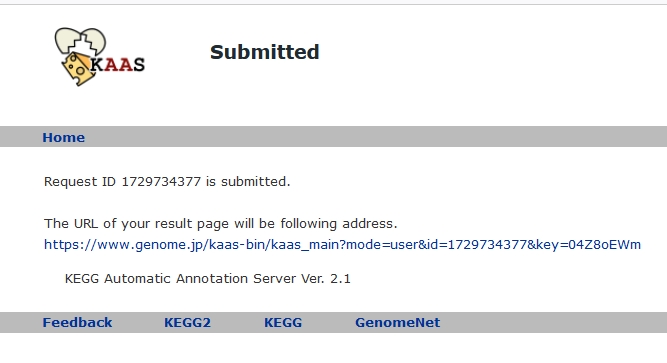
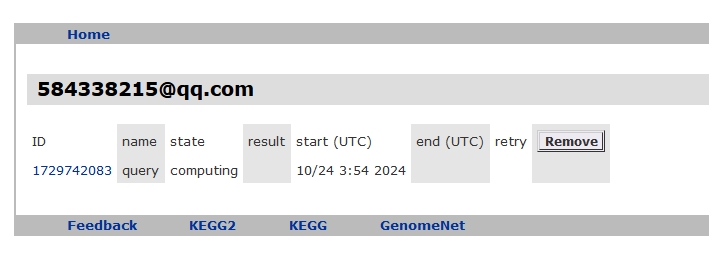
Wait for the completion of the job.
Quote
Email me with more questions! 584338215@qq.com
